Public Data Access Overview / Example Scripts
We provide example scripts for reading the data files of the Illustris[TNG] simulations. They are available in a few languages, with identical functionality in each. This includes:
- (i) reading a given particle type and/or data field from the snapshot files,
- (ii) reading only the particle subset from the snapshot corresponding to a halo or subhalo,
- (iii) extracting the full subtree or main progenitor branch from either SubLink or LHaloTree for a given subhalo,
- (iv) walking a tree to count the number of mergers,
- (v) reading the entire group catalog at one snapshot,
- (vi) reading specific fields from the group catalog, or the entries for a single halo or subhalo.
We expect they will provide a useful starting point for writing any analysis task, and intend them as a 'minimal working examples' which are short and simple enough that they can be quickly understood and extended.
Currently available are: Python (3.6+ recommended), IDL (8.0+ required), Matlab (R2013a+ required), and Julia (1.6+ recommended). Select one to show all the content on this page specifically for that language.
In all cases, these scripts assume that you have downloaded local copies of the relevant files.
Paths to files are defined with respect to a basePath which is
passed in to all read functions. The locations of group catalog files, snapshot files, and
merger trees files are then specified in a handful of functions, e.g. gcPath(),
snapPath(), and treePath(). These can be modified as necessary to
point to your local files, but it is best to keep a directory structure organized as e.g.:
TNG100-1/TNG100-1/output/- group catalogs:
TNG100-1/output/groups_099/fof_subhalo_tab_099.*.hdf5 - snapshots:
TNG100-1/output/snapdir_099/snap_099.*.hdf5 TNG100-1/postprocessing/- offsets:
TNG100-1/postprocessing/offsets/offsets_*.hdf5 - SubLink mergertree:
TNG100-1/postprocessing/trees/SubLink/tree_extended.*.hdf5 - other catalogs:
TNG100-1/postprocessing/catalog_name/files*.hdf5
where you would set basePath = 'TNG100-1/output/' in the following.
Getting Started Guide
We walk through the process of downloading and exploring data in the Subfind group catalogs, raw snapshot files, and the SubLink merger tree. In these examples we will work with Illustris-3 since the file sizes are smaller to download and easier to work with, although you can replace each occurrence with e.g. 'Illustris-1', 'TNG100-1', or 'TNG300-1' for the flagship simulations.
If you haven't already, let's download the example scripts from Github and make sure they installed (Python and Julia) or on the appropriate path for loading (Matlab and IDL). Specifically:
$ cd ~
$ git clone https://github.com/illustristng/illustris_python.git
$ pip install illustris_python/
$ cd ~
$ git clone https://github.com/illustristng/illustris_idl.git
$ export IDL_PATH=~/illustris_idl # add to .bashrc
$ cd ~
$ git clone https://github.com/illustristng/illustris_matlab.git
$ export MATLABPATH=~/illustris_matlab # add to .bashrc
julia> press "]"
pkg> add https://github.com/illustristng/illustris_julia.git
Group Catalogs
First, make a base directory for the run and a subdirectory for the z=0 group
catalogs (snapshot 135), then download the catalog (~100 MB).
$ mkdir -p ~/Illustris-3/output/groups_135
$ cd ~/Illustris-3/output/groups_135/
$ wget -nd -nc -nv -e robots=off -l 1 -r -A hdf5 --content-disposition --header="API-Key: INSERT_API_KEY_HERE" "http://www.tng-project.org/api/Illustris-3/files/groupcat-135/?format=api"
Note: Separate offset files for TNG
For all TNG simulations the "offset" files are separate and must be downloaded in addition to the group catalog files, in order to load (i) single halos or subhalos, or (ii) load the particles of particular halos or subhalos. For example, if you were working with TNG100-3 at z=0, then:$ mkdir -p ~/TNG100-3/postprocessing/offsets/
$ cd ~/TNG100-3/postprocessing/offsets/
$ wget --content-disposition --header="API-Key: INSERT_API_KEY_HERE" "http://www.tng-project.org/api/TNG100-3/files/offsets.99.hdf5"
Start up your interface of choice and import the example scripts:
$ python
>>> import illustris_python as il
>>>
$ idl
IDL> .r illustris
% Compiled module: PARTTYPENUM
% Compiled module: (many more)
IDL>
$ matlab
>>
$ julia
julia> import illustris_julia as il
[ Info: Precompiling illustris_julia [356b6c62-7396-11ec-31d4-55c52b5be5ca]
julia>
Define the base path for the data (modify as needed), and load the total masses (SubhaloMass) and star formation rate within twice the stellar half mass radius (SubhaloSFRinRad) of all the Subfind subhalos.
>>> basePath = './Illustris-3/output/'
>>> fields = ['SubhaloMass','SubhaloSFRinRad']
>>> subhalos = il.groupcat.loadSubhalos(basePath,135,fields=fields)
IDL> basePath = "./Illustris-3/output/"
IDL> fields = ['SubhaloMass','SubhaloSFRinRad']
IDL> subhalos = loadSubhalos(basePath,135,fields=fields)
>> basePath = './Illustris-3/output/';
>> fields = {'SubhaloMass','SubhaloSFRinRad'};
>> subhalos = illustris.groupcat.loadSubhalos(basePath,135,fields);
julia> basePath = "./Illustris-3/output/";
julia> fields = ["SubhaloMass","SubhaloSFRinRad"];
julia> subhalos = il.groupcat.loadSubhalos(basePath,135,fields);
Inspecting the return, we see it is a dict/hash/struct with a key count
indicating that there are 121209 total subhalos. Each requested field is returned
as a numeric array with a key name equal to its field name in the group catalog.
>>> subhalos.keys()
['count', 'SubhaloSFRinRad', 'SubhaloMass']
>>> subhalos['SubhaloMass'].shape
(121209,)
IDL> print, subhalos.keys()
count
SubhaloMass
SubhaloSFRinRad
IDL> help, subhalos['SubhaloMass']
<Expression> FLOAT = Array[121209]
>> subhalos
subhalos =
count: 121209
SubhaloMass: [1x121209 single]
SubhaloSFRinRad: [1x121209 single]
julia> subhalos
Dict{Any, Any} with 3 entries:
"SubhaloMass" => Float32[25250.1, 2467.93, 406.137, 310.241, …
"count" => 121209
"SubhaloSFRinRad" => Float32[4.11098, 1.87522, 0.344028, 4.04084, …
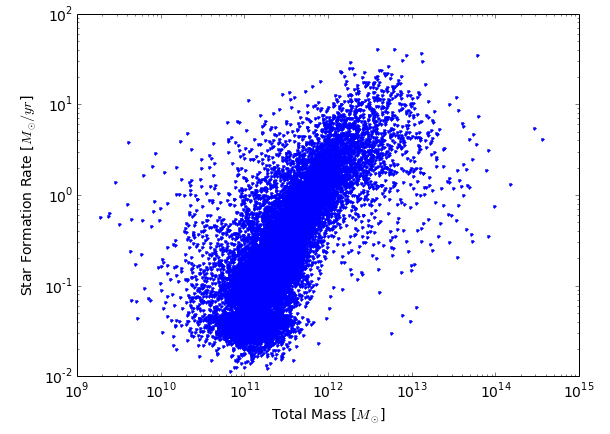
Make a simple scatterplot of the relation (note the units).
>>> import matplotlib.pyplot as plt
>>> mass_msun = subhalos['SubhaloMass'] * 1e10 / 0.704
>>> plt.plot(mass_msun,subhalos['SubhaloSFRinRad'],'.')
>>> plt.xscale('log')
>>> plt.yscale('log')
>>> plt.xlabel('Total Mass [$M_\odot$]')
>>> plt.ylabel('Star Formation Rate [$M_\odot / yr$]')
IDL> mass_msun = subhalos['SubhaloMass'] * 1e10 / 0.704
IDL> p = plot(mass_msun, subhalos['SubhaloSFRinRad'], '.', /xlog, /ylog)
IDL> p.xtitle = "Total Mass [$M_{\rm sun}$]"
IDL> p.ytitle = "Star Formation Rate [$M_{\rm sun}$/yr]"
>> mass_msun = subhalos.('SubhaloMass') * 1e10 / 0.704;
>> loglog(mass_msun, subhalos.('SubhaloSFRinRad'), '.');
>> xlabel('Total Mass [$M_\odot$]');
>> ylabel('Star Formation Rate [$M_\odot / yr$]');
julia> using Plots
julia> mass_msun = subhalos["SubhaloMass"] * 1e10 / 0.704;
julia> scatter(mass_msun, subhalos["SubhaloSFRinRad"], xaxis=:log, yaxis=:log, ylims=[1e-2,1e2]);
julia> scatter!(xlabel="Total Mass [Msun]", ylabel="Star Formation Rate [Msun/yr]")
# to save the Figure to a PDF file without needing an interactive display window:
julia> ENV["GKSwstype"]="nul"; # after 'using Plots'
julia> savefig("out.pdf"); # after all plotting commands
Let us get a list of primary subhalo IDs by loading the GroupFirstSub
field from the FoF groups.
>>> GroupFirstSub = il.groupcat.loadHalos(basePath,135,fields=['GroupFirstSub'])
>>> GroupFirstSub.dtype
dtype('uint32')
>>> GroupFirstSub.shape
(131727,)
IDL> GroupFirstSub = loadHalos(basePath,135,fields=['GroupFirstSub'])
IDL> help, GroupFirstSub
GROUPFIRSTSUB ULONG = Array[131727]
>> GroupFirstSub = illustris.groupcat.loadHalos(basePath,135,{'GroupFirstSub'});
>> class(GroupFirstSub)
ans = uint32
>> size(GroupFirstSub)
ans = 1 131727
julia> GroupFirstSub = il.groupcat.loadHalos(basePath,135,["GroupFirstSub"]);
julia> eltype(GroupFirstSub)
UInt32
julia> size(GroupFirstSub)
(131727,)
Note: Return type
When a single field is requested, the default return is a raw numeric array and not a container type object.For the 5 most massive central subhalos, let's load all their fields from the group catalog and print a gas fraction (gas mass over total baryonic mass) in the stellar half mass radius.
>>> ptNumGas = il.snapshot.partTypeNum('gas') # 0
>>> ptNumStars = il.snapshot.partTypeNum('stars') # 4
>>> for i in range(5):
>>> all_fields = il.groupcat.loadSingle(basePath,135,subhaloID=GroupFirstSub[i])
>>> gas_mass = all_fields['SubhaloMassInHalfRadType'][ptNumGas]
>>> stars_mass = all_fields['SubhaloMassInHalfRadType'][ptNumStars]
>>> frac = gas_mass / (gas_mass + stars_mass)
>>> print GroupFirstSub[i], frac
0 0.0688846
608 0.0236937
1030 0.0638515
1396 0.00357705
1801 0.1222
IDL> ptNumGas = partTypeNum('gas') ; 0
IDL> ptNumStars = partTypeNum('stars') ; 4
IDL> for i=0,4 do begin
IDL> all_fields = loadGroupcatSingle(basePath,135,subhaloID=GroupFirstSub[i])
IDL> gas_mass = all_fields['SubhaloMassInHalfRadType',ptNumGas]
IDL> stars_mass = all_fields['SubhaloMassInHalfRadType',ptNumStars]
IDL> frac = gas_mass / (gas_mass + stars_mass)
IDL> print, GroupFirstSub[i], frac
IDL> endfor
0 0.0688846
608 0.0236937
1030 0.0638515
1396 0.00357705
1801 0.122200
>> ptNumGas = illustris.partTypeNum('gas'); % 0
>> ptNumStars = illustris.partTypeNum('stars'); % 4
>> for i = 1:5
>> all_fields = illustris.groupcat.loadSingle(basePath,135,'subhalo',GroupFirstSub(i));
>> % take care with particle type numbers and 1-based indexing
>> gas_mass = all_fields.('SubhaloMassInHalfRadType')(ptNumGas+1);
>> stars_mass = all_fields.('SubhaloMassInHalfRadType')(ptNumStars+1);
>> frac = gas_mass / (gas_mass + stars_mass);
>> fprintf('%d %f\n', GroupFirstSub(i), frac);
>> end
0 0.068885
608 0.023694
1030 0.063851
1396 0.003577
1801 0.122200
julia> ptNumGas = il.util.partTypeNum("gas");
julia> ptNumStars = il.util.partTypeNum("stars");
julia> for i = 1:5
all_fields = il.groupcat.loadSingle(basePath,135,subhaloID=GroupFirstSub[i]);
gas_mass = all_fields["SubhaloMassInHalfRadType"][ptNumGas+1];
stars_mass = all_fields["SubhaloMassInHalfRadType"][ptNumStars+1];
frac = gas_mass / (gas_mass + stars_mass);
print(GroupFirstSub[i], ' ', frac, '\n')
end
0 0.06888455
608 0.023693658
1030 0.06385147
1396 0.003577049
1801 0.12220047
Merger Trees
Let's download the full SubLink merger tree to play with (~8 GB).
$ mkdir -p ~/Illustris-3/postprocessing/trees/SubLink
$ cd ~/Illustris-3/postprocessing/trees/SubLink/
$ wget -nd -nc -nv -e robots=off -l 1 -r -A hdf5 --content-disposition --header="API-Key: INSERT_API_KEY_HERE" "http://www.tng-project.org/api/Illustris-3/files/sublink/?format=api"
Reading individual sub-trees from the full merger trees with 'offsets'
The example scripts make use of pre-computed offsets to accelerate the reading of subsets of the full merger tree files. For example, in order to load the main progenitor branch of the SubLink tree for a single subhalo. These offsets are stored in the group catalog files. Therefore, theloadTree() function requires that you have already downloaded the group catalogs
corresponding to the snapshot of the subhalo (in addition to the tree files themselves).
For the 101st through 105th (indices 100 through 104 if 0-based) most massive primaries, extract the total mass, Subfind ID, and snapshot along the main progenitor branch. Plot the mass histories.
>>> fields = ['SubhaloMass','SubfindID','SnapNum']
>>> start = 100
>>> for i in range(start,start+5):
>>> tree = il.sublink.loadTree(basePath,135,GroupFirstSub[i],fields=fields,onlyMPB=True)
>>> plt.plot(tree['SnapNum'],tree['SubhaloMass'],'-')
>>> plt.yscale('log')
>>> plt.xlabel('Snapshot Number')
>>> plt.ylabel('Total Subhalo Mass [code units]')
IDL> fields = ['SubhaloMass','SubfindID','SnapNum']
IDL> start = 100
IDL> for i=start,start+4 do begin
IDL> tree = loadSublinkTree(basePath,135,GroupFirstSub[i],fields=fields,/onlyMPB)
IDL> p = plot(tree['SnapNum'],tree['SubhaloMass'],'-',/ylog,overplot=(i gt start))
IDL> p.color = (['b','g','r','c','m'])(i-start)
IDL> endfor
IDL> p.xtitle = "Snapshot Number"
IDL> p.ytitle = "Total Subhalo Mass [code units]"
>> fields = {'SubhaloMass','SubfindID','SnapNum'};
>> start = 100;
>> for i = start+1:start+5 % take care with 1-based indexing!
>> tree = illustris.sublink.loadTree(basePath,135,GroupFirstSub(i),fields,true);
>> semilogy(tree.('SnapNum'), tree.('SubhaloMass'), '-');
>> end
>> xlabel('Total Mass [$M_\odot$]');
>> ylabel('Star Formation Rate [$M_\odot$ / yr]');
julia> fields = ["SubhaloMass","SubfindID","SnapNum"];
julia> start = 100;
julia> for i = start+1:start+5 # take care with 1-based indexing!
tree = il.sublink.loadTree(basePath,135,GroupFirstSub[i],fields,true);
plot!(tree["SnapNum"], tree["SubhaloMass"], yaxis=:log);
end
julia> plot!(xlabel="Snapshot Number");
julia> plot!(ylabel="Total Subhalo Mass [code units]");
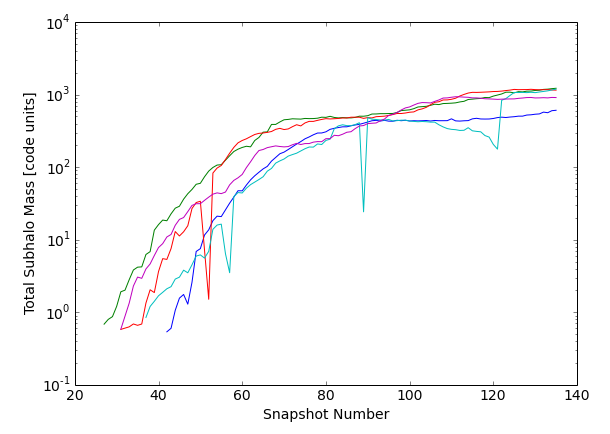
Note that the single-snapshot dips seen in the cyan and red curves can sometimes occur due to the 'subhalo switching problem'. The downward trend in mass followed by the sudden increase in the cyan is a signature of a merger. For details on both aspects of the trees, see the SubLink paper.
We include a semi-complex example of walking through the tree to determine the number of past mergers of a given subhalo, above some mass ratio threshold. Here, the mass ratio is defined as the ratio of the maximum past stellar mass of the two progenitors. For the same halos as above, count the number of major mergers (mass ratio > 1/5).
>>> ratio = 1.0/5.0
>>> # the following fields are required for the walk and the mass ratio analysis
>>> fields = ['SubhaloID','NextProgenitorID','MainLeafProgenitorID','FirstProgenitorID','SubhaloMassType']
>>> for i in range(start,start+5):
>>> tree = il.sublink.loadTree(basePath,135,GroupFirstSub[i],fields=fields)
>>> numMergers = il.sublink.numMergers(tree,minMassRatio=ratio)
>>> print GroupFirstSub[i], numMergers
9106 4
9137 2
9151 3
9170 5
9191 2
IDL> ratio = 1.0/5.0
IDL> ; the following fields are required for the walk and the mass ratio analysis
IDL> fields = ['SubhaloID','NextProgenitorID','MainLeafProgenitorID','FirstProgenitorID','SubhaloMassType']
IDL> for i=start,start+4 do begin
IDL> tree = loadSublinkTree(basePath,135,GroupFirstSub[i],fields=fields)
IDL> numMergers = numMergers(tree,minMassRatio=ratio)
IDL> print, GroupFirstSub[i], numMergers
IDL> endfor
9106 4
9137 2
9151 3
9170 5
9191 2
>> ratio = 1.0/5.0;
>> % the following fields are required for the walk and the mass ratio analysis
>> fields = {'SubhaloID','NextProgenitorID','MainLeafProgenitorID','FirstProgenitorID','SubhaloMassType'};
>> for i = start+1:start+5
>> tree = illustris.sublink.loadTree(basePath,135,GroupFirstSub(i),fields);
>> numMergers = illustris.sublink.numMergers(tree,ratio);
>> fprintf('%d %d\n', GroupFirstSub(i), numMergers);
>> end
9106 4
9137 2
9151 3
9170 5
9191 2
julia> ratio = 1/5;
julia> # the following fields are required for the walk and the mass ratio analysis
julia> fields = ["SubhaloID","NextProgenitorID","MainLeafProgenitorID","FirstProgenitorID","SubhaloMassType"];
julia> for i = start+1:start+5
tree = il.sublink.loadTree(basePath,135,GroupFirstSub[i],fields);
numMergers = il.sublink.numMergers(tree,ratio);
print(GroupFirstSub[i], " ", numMergers, "\n");
end
9106 4
9137 2
9151 3
9170 5
9191 2
Snapshot Data
Let's download the full z=0 snapshot to play with (~20 GB).
$ mkdir ~/Illustris-3/output/snapdir_135
$ cd ~/Illustris-3/output/snapdir_135/
$ wget -nd -nc -nv -e robots=off -l 1 -r -A hdf5 --content-disposition --header="API-Key: INSERT_API_KEY_HERE" "http://www.tng-project.org/api/Illustris-3/files/snapshot-135/?format=api"
Reading individual halos and subhalos from the snapshots with 'offsets'
As with the merger trees, reading subsets of the snapshots makes use of pre-computed offsets. For example, in order to load only the particles belonging to a single FoF halo. For Illustris, these offsets are stored in the group catalog files. For TNG, these offsets are stored in separate "offset" files, as described above. As a result, theloadSubhalo() and loadHalo() functions require that
you have already downloaded the corresponding offset files (in addition to the snapshot files
themselves).
First, load the Masses of all the gas cells in the entire box, and calculate
their mean, converting to log solar masses.
>>> import numpy as np
>>> fields = ['Masses']
>>> gas_mass = il.snapshot.loadSubset(basePath,135,'gas',fields=fields)
>>> print np.log10( np.mean(gas_mass,dtype='double')*1e10/0.704 )
7.92160161941
IDL> fields = ['Masses']
IDL> gas_mass = loadSnapSubset(basePath,135,'gas',fields=fields)
IDL> print, alog10( mean(gas_mass)*1e10/0.704 )
7.91676
>> fields = {'Masses'};
>> gas_mass = illustris.snapshot.loadSubset(basePath,135,'gas',fields);
>> gas_mass_mean = sum(gas_mass,'double')/numel(gas_mass); % mean() has significant float error
>> disp( log10( gas_mass_mean*1e10/0.704 ))
7.9216
julia> using Statistics
julia> fields = ["Masses"];
julia> gas_mass = il.snapshot.loadSubset(basePath,135,"gas",fields);
julia> gas_mass_mean = log10( mean(gas_mass)*1e10/0.704 )
7.921601668256946
Next, load the spatial Coordinates of all the dark matter particles in the box,
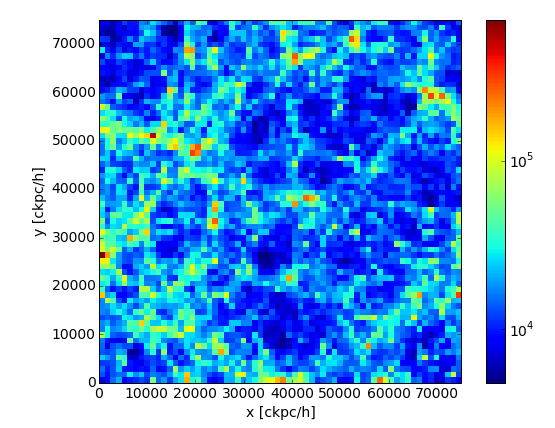
and make a quick image with a 2D histogram, projecting out the z-axis.
>>> import matplotlib as mpl
>>> dm_pos = il.snapshot.loadSubset(basePath,135,'dm',['Coordinates']);
>>> plt.hist2d(dm_pos[:,0], dm_pos[:,1], norm=mpl.colors.LogNorm(), bins=64);
>>> plt.xlim([0,75000])
>>> plt.ylim([0,75000])
>>> plt.xlabel('x [ckpc/h]')
>>> plt.ylabel('y [ckpc/h]')
IDL> dm_pos = loadSnapshotSubset(basePath,135,'dm',fields=['Coordinates']);
IDL> bin = 75000.0/64
IDL> h = hist_2d(dm_pos[0,*], dm_pos[1,*], bin1=bin,bin2=bin,min1=0,min2=0,max1=75000,max2=75000)
IDL> tv, bytscl(alog10(h>1)), xs=10, ys=10
>> dm_pos = illustris.snapshot.loadSubset(basePath,135,'dm',{'Coordinates'});
>> h = hist3([dm_pos(1,:)' dm_pos(2,:)'], [64 64]);
>> imagesc(log10(h));
julia> dm_pos = il.snapshot.loadSubset(basePath,135,"dm",["Coordinates"]);
julia> histogram2d(dm_pos[1,:], dm_pos[2,:], colorbar_scale = :log10, nbins=64)
julia> histogram2d!(xlim=[0,75000], ylim=[0,75000], xlabel="x [ckpc/h]", ylabel="y [ckpc/h]")
Finally, load the star particles belonging to FoF halo ID 100 (all fields). Print the minimum and maximum of all positions for each axis to check we have loaded only stars in a localized region.
>>> stars = il.snapshot.loadHalo(basePath,135,100,'stars')
>>> stars.keys()
['count', u'GFM_Metals', u'SubfindVelDisp', u'GFM_InitialMass', u'Masses', u'Velocities', u'Coordinates', u'Potential', u'SubfindHsml', u'SubfindDensity', u'NumTracers', u'ParticleIDs', u'GFM_StellarFormationTime', u'GFM_StellarPhotometrics', u'GFM_Metallicity']
>>> for i in range(3):
>>> print np.min(stars['Coordinates'][:,i]), np.max(stars['Coordinates'][:,i])
17993.7 19585.6
58373.0 59606.2
67596.5 68610.6
IDL> stars = loadHalo(basePath,135,100,'stars')
IDL> print, stars.keys()
Masses
GFM_InitialMass
Potential
SubfindHsml
Velocities
Coordinates
count
GFM_StellarFormationTime
GFM_Metallicity
SubfindDensity
GFM_Metals
NumTracers
SubfindVelDisp
GFM_StellarPhotometrics
ParticleIDs
IDL> for i=0,2 do print, minmax( (stars['Coordinates'])[i,*] )
17993.7 19585.6
58373.0 59606.2
67596.5 68610.6
>> stars = illustris.snapshot.loadHalo(basePath,135,100,'stars');
>> stars
stars =
count: 5548
Coordinates: [3x5548 single]
GFM_InitialMass: [1x5548 single]
GFM_Metallicity: [1x5548 single]
GFM_Metals: [9x5548 single]
GFM_StellarFormationTime: [1x5548 single]
GFM_StellarPhotometrics: [8x5548 single]
Masses: [1x5548 single]
NumTracers: [1x5548 uint32]
ParticleIDs: [1x5548 uint64]
Potential: [1x5548 single]
SubfindDensity: [1x5548 single]
SubfindHsml: [1x5548 single]
SubfindVelDisp: [1x5548 single]
Velocities: [3x5548 single]
>> for i = 1:3
>> fprintf('%g %g\n', min(stars.('Coordinates')(i,:)), max(stars.('Coordinates')(i,:)))
>> end
17993.7 19585.6
58373 59606.2
67596.5 68610.6
julia> stars = il.snapshot.loadHalo(basePath,135,100,"stars")
Dict{Any, Any} with 16 entries:
"Masses" => Float32[0.00617214, 0.0047529, 0.005002, …
"NumTracers" => UInt32[0x00000000, 0x00000000, 0x00000000, …
"Potential" => Float32[-8.18708f5, -8.18263f5, -8.17523f5, …
"ParticleIDs" => UInt64[0x0000001758c9f2c3, 0x000000175472a1d5, 0x000000175b468ce8, …
"GFM_InitialMass" => Float32[0.011336, 0.00872101, 0.00911615, …
"count" => 0x000015ac
"StellarHsml" => Float32[1.08076, 1.10283, 1.13628, …
"GFM_Metallicity" => Float32[0.0370382, 0.0168663, 0.0532344, …
"GFM_Metals" => Float32[0.706836 0.733932 …
"SubfindDensity" => Float32[0.041891, 0.0415178, …
"SubfindVelDisp" => Float32[265.83, 270.626, 262.545, …
"Velocities" => Float32[-110.249 -108.5 …
"SubfindHsml" => Float32[3.367, 3.367, 3.57803, …
"GFM_StellarPhotometrics" => Float32[-11.93 -11.935 …
"Coordinates" => Float32[18314.2 18314.1 …
"GFM_StellarFormationTime" => Float32[0.503055, 0.381383, 0.619853, …
julia> for i in 1:3
print(minimum(stars["Coordinates"][i,:]), " ", maximum(stars["Coordinates"][i,:]), "\n");
end
17993.686 19585.623
58372.957 59606.2
67596.48 68610.61
That's it! This should give you a good starting point, together with the reference below, to any aspect of the simulation data.
scida - scalable analysis for scientific big data
Interested in working with TNG data using a higher-level package, that automatically handles many complex tasks such as data loading, units, parallel calculations, and more?Check out scida, which has built-in support for Illustris, TNG, EAGLE, SIMBA, and many other cosmological hydrodynamical simulations.
I/O Scripts Reference
Loading from the FoF and Subfind group catalogs:
The optional fields argument always accepts a string list/array of field names,
which must agree (case-sensitive) to the available datasets in the group catalog. If it is not
specified, all fields will be read and returned, which could be significantly slower.
illustris_python.groupcat.
def loadSubhalos(basePath, snapNum, fields=None):
""" Load all subhalo information from the entire group catalog for one snapshot
(optionally restrict to a subset given by fields). """
def loadHalos(basePath, snapNum, fields=None):
""" Load all halo information from the entire group catalog for one snapshot
(optionally restrict to a subset given by fields). """
def loadHeader(basePath, snapNum):
""" Load the group catalog header. """
def load(basePath, snapNum):
""" Load complete group catalog all at once. """
def loadSingle(basePath, snapNum, haloID=-1, subhaloID=-1):
""" Return complete group catalog information for one halo or subhalo. """
function loadSubhalos(basePath, snapNum, fields=fields)
; Load all subhalo information from the entire group catalog for one snapshot
; (optionally restrict to a subset given by fields).
function loadHalos(basePath, snapNum, fields=fields)
; Load all halo information from the entire group catalog for one snapshot
; (optionally restrict to a subset given by fields).
function loadHeader(basePath, snapNum, chunkNum=chunkNum)
; Load the group catalog header (chunkNum=0 if not specified).
function loadGroupcat(basePath, snapNum)
; Load complete group catalog all at once.
function loadGroupcatSingle(basePath, snapNum, haloID=hID, subhaloID=shID)
; Return complete group catalog information for one halo or subhalo.
illustris.groupcat.
function [result] = loadSubhalos(basePath, snapNum, fields)
% LOADSUBHALOS Load all subhalo information from the entire group catalog for one snapshot
% (optionally restrict to a subset given by fields).
function [result] = loadHalos(basePath, snapNum, fields)
% LOADHALOS Load all halo information from the entire group catalog for one snapshot
% (optionally restrict to a subset given by fields).
function [header] = loadHeader(basePath, snapNum, chunkNum)
% LOADHEADER Load the group catalog header (chunkNum=0 if not specified).
function [gc] = load(basePath, snapNum)
% LOAD Load complete group catalog all at once.
function [result] = loadSingle(basePath, snapNum, type, id)
% LOADSINGLE Return complete group catalog information for one halo or subhalo.
% Type should be one of {'halo','group','subhalo','subgroup'}.
illustris_julia.groupcat.
"Load all subhalo information from the entire group catalog for one snapshot (optionally restrict to a subset given by fields)."
function loadSubhalos(basePath, snapNum, fields=nothing)
"Load all halo information from the entire group catalog for one snapshot (optionally restrict to a subset given by fields)."
function loadHalos(basePath, snapNum, fields=nothing)
"Load the group catalog header."
function loadHeader(basePath, snapNum)
"Load complete group catalog all at once."
function load(basePath, snapNum)
"Return complete group catalog information for one halo or subhalo."
function loadSingle(basePath, snapNum; haloID=-1, subhaloID=-1)
Loading particle-level data from the snapshots:
The optional fields argument always accepts a string list/array of field names,
which must agree (case-sensitive) to the available datasets for that particle type in the
snapshot. If it is not specified, all fields will be read and returned, which could be significantly slower.
The partType argument may either be the particle type number, or one of the recognized
string names, e.g. 'gas', 'stars', 'bhs', or 'dm'.
illustris_python.snapshot.
def loadSubset(basePath, snapNum, partType, fields=None):
""" Load a subset of fields for all particles/cells of a given partType. """
def loadSubhalo(basePath, snapNum, id, partType, fields=None):
""" Load all particles/cells of one type for a specific subhalo
(optionally restricted to a subset fields). """
def loadHalo(basePath, snapNum, id, partType, fields=None):
""" Load all particles/cells of one type for a specific halo
(optionally restricted to a subset fields). """
function loadSnapSubset(basePath, snapNum, partType, fields=fields)
; Load a subset of fields for all particles/cells of a given partType.
function loadSubhalo(basePath, snapNum, id, partType, fields=fields)
; Load all particles/cells of one type for a specific subhalo
; (optionally restricted to a subset fields).
function loadHalo(basePath, snapNum, id, partType, fields=fields)
; Load all particles/cells of one type for a specific halo
; (optionally restricted to a subset fields).
illustris.snapshot.
function [result] = loadSubset(basePath, snapNum, partType, fields)
% LOADSUBET Load a subset of fields for all particles/cells of a given partType.
function [result] = loadSubhalo(basePath, snapNum, id, partType, fields)
% LOADSUBHALO Load all particles/cells of one type for a specific subhalo
% (optionally restricted to a subset fields).
function [result] = loadHalo(basePath, snapNum, id, partType, fields)
% LOADHALO Load all particles/cells of one type for a specific halo
% (optionally restricted to a subset fields).
illustris_julia.snapshot.
"Load a subset of fields for all particles/cells of a given partType."
function loadSubset(basePath, snapNum, partType, fields=nothing)
"Load all particles/cells of one type for a specific subhalo (optionally restricted to a subset fields)."
function loadSubhalo(basePath, snapNum, id, partType, fields=nothing)
"Load all particles/cells of one type for a specific halo (optionally restricted to a subset fields)."
function loadHalo(basePath, snapNum, id, partType, fields=nothing)
Loading from the SubLink merger trees:
The optional fields argument always accepts a string list/array of field names,
which must agree (case-sensitive) to the available datasets in the SubLink trees.
If it is not specified, all fields will be read and returned, which could be significantly slower.
If onlyMPB = True, then only the main progenitor branch will be loaded (that is,
only FirstProgenitor links will be followed).
illustris_python.sublink.
def loadTree(basePath, snapNum, id, fields=None, onlyMPB=False, onlyMDB=False):
""" Load portion of Sublink tree, for a given subhalo, in its existing flat format.
(optionally restricted to a subset fields). If onlyMPB, then only the 'main progenitor branch'
is loaded. If onlyMDB, then only the 'main descendant branch' is loaded, which is a single
tree branch if and only if this subhalo lies on the MPB of its z=0 descendant, while if not,
then the return is the full descendant merger tree which contains as its first entry the z=0
descendant of this subhalo. """
def numMergers(tree, minMassRatio=1e-10, massPartType='stars', index=0):
""" Calculate the number of mergers, along the main-progenitor branch, in this sub-tree (optionally above some mass ratio threshold). """
function loadSublinkTree(basePath, snapNum, id, fields=fields, onlyMPB=onlyMPB)
; Load portion of Sublink tree, for a given subhalo, in its existing flat format.
; (optionally restricted to a subset fields).
function numMergers(tree, minMassRatio=1e-10, massPartType='stars', index=0)
; Calculate the number of mergers, along the main-progenitor branch, in this sub-tree (optionally above some mass ratio threshold).
illustris.sublink.
function [result] = loadTree(basePath, snapNum, id, fields, onlyMPB)
% LOADTREE Load portion of Sublink tree, for a given subhalo, in its existing flat format.
% (optionally restricted to a subset fields).
function [numMergers] = numMergers(tree, minMassRatio, massPartType, index)
% NUMMERGERS Calculate the number of mergers, along the main-progenitor branch, in this sub-tree
% (optionally above some mass ratio threshold).
illustris_julia.sublink.
"Load portion of Sublink tree, for a given subhalo, in its existing flat format (optionally restricted to a subset fields)."
function loadTree(basePath, snapNum, id, fields=nothing, onlyMPB=false)
"Calculate the number of mergers in this sub-tree (optionally above some mass ratio threshold)."
function numMergers(tree, minMassRatio=1e-10, massPartType="stars", index=0)
Loading from the LHaloTree merger trees:
The optional fields argument always accepts a string list/array of field names,
which must agree (case-sensitive) to the available datasets in the LHaloTree trees.
If it is not specified, all fields will be read and returned, which could be significantly slower.
If onlyMPB = True, then only the main progenitor branch will be loaded (that is,
only FirstProgenitor links will be followed).
illustris_python.lhalotree.
def loadTree(basePath, snapNum, id, fields=None, onlyMPB=False):
""" Load portion of LHaloTree, for a given subhalo, re-arranging into a flat format. """
function loadLHaloTree(basePath, snapNum, id, fields=fields, onlyMPB=onlyMPB)
; Load portion of LHaloTree, for a given subhalo, re-arranging into a flat format.
illustris.lhalotree.
function [result] = loadTree(basePath,snapNum,id,fields,onlyMPB)
% LOADTREE Load portion of LHaloTree tree, for a given subhalo, re-arranging into a flat format.
illustris_julia.lhalotree.
"Load portion of LHaloTree, for a given subhalo, re-arranging into a flat format."
function loadTree(basePath, snapNum, id, fields=nothing, onlyMPB=false)
General utility functions:
illustris_python.util.
def partTypeNum(partType):
""" Mapping between common names and numeric particle types. """
function partTypeNum(PT)
; Mapping between common names and numeric particle types.
illustris.
function [s] = partTypeNum(pt)
% PARTTYPENUM Mapping between common names and numeric particle types.
illustris_julia.util.
"Mapping between common names and numeric particle types."
function partTypeNum(partType)